Post-Translational modifications of proteins and DNA repair: structure, function & dynamic
- Decoding the mechanism of protein SUMOylation and related applications
Protein SUMOylation (conjugation of the small SUMO protein to the side chain of a lysine) is an essential post-translational modification (PTM) in eukaryotes. One third of human proteins are substrates of SUMOylation.
At the cellular level, it plays a fundamental role in transcriptional repression and the response to various stress factors, including DNA damage. For example, dysregulation of SUMOylation confers increased resistance to anti-tumour treatments in certain cancer cells.
From an evolutionary perspective, SUMOylation is related to ubiquitination but, unlike the latter, remains poorly understood and still poorly documented. Both modifications are introduced by an enzymatic cascade involving three types of ligase, called E1, E2 and E3. E3 ligases play a key role in the specificity of the cascade for a given substrate. While more than 600 human ubiquitin E3 ligases are known, only about 15 equivalent enzymes are known for SUMO. The functional consequences of SUMOylation are also much less clear than those of ubiquitination. While the latter mainly promotes protein degradation, SUMOylation is thought to play more subtle regulatory roles by inducing or blocking macromolecular interactions. However, for most substrates, the effect of SUMOylation is unknown.
- Molecular and atomic-level deciphering of how DNA damage is recognised and eliminated by enzymes in the base excision repair (BER) system
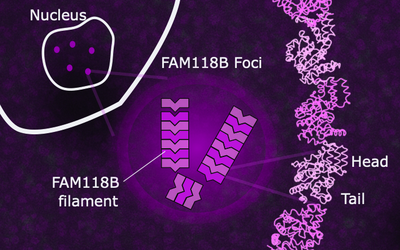
FAM118B and FAM118A : new sirtuins of bacterial origin, filament, in the heart of human cells
Published on Jan. 28, 2026


Aranyadip Gayen seminar
Published on Sept. 12, 2025
