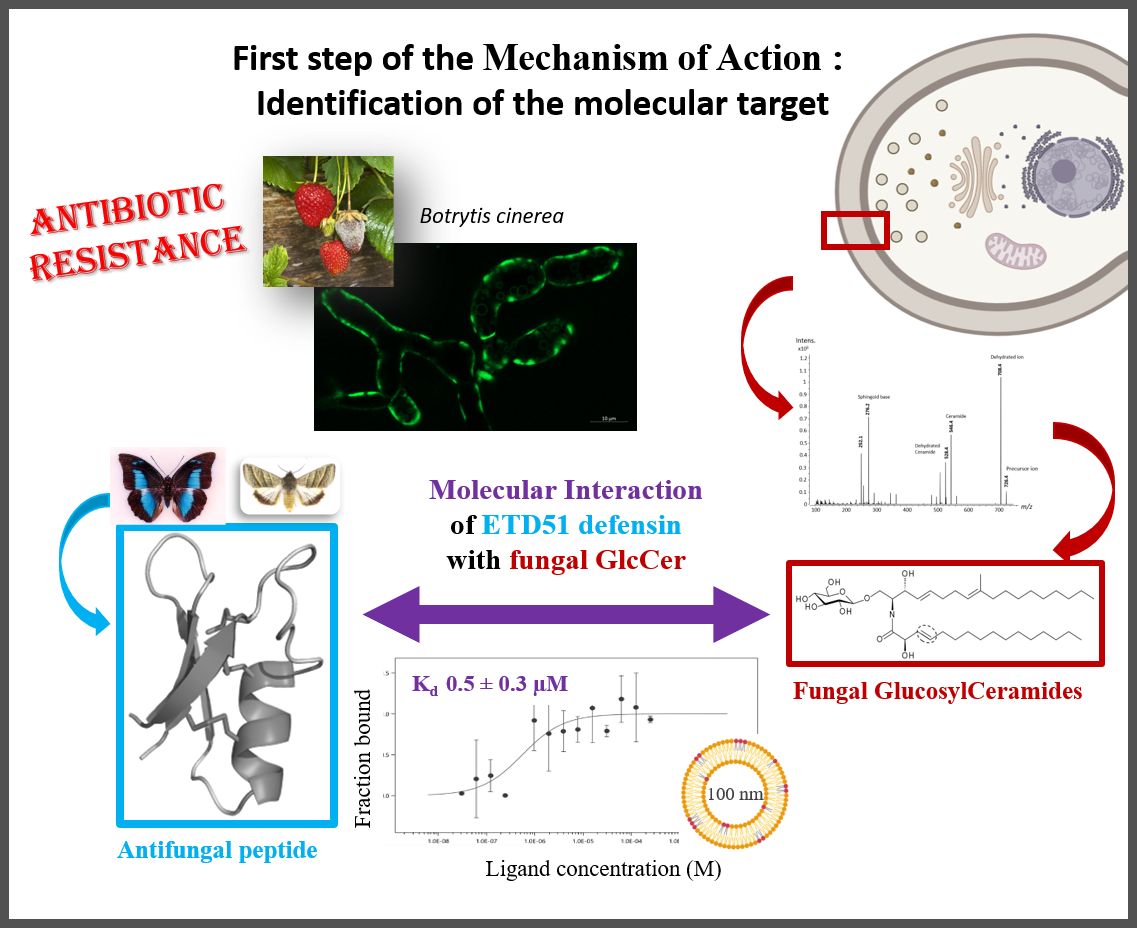
Researchers from the CBM's “NMR of Biomolecules” team have identified the molecular target of ETD151, an antifungal peptide optimized from butterfly defensins. Active against a range of pathogenic fungi, this antifungal peptide thus opens up its full potential in both medicine and agriculture.
A lire aussi
6 novembre 2025 - Soutenance de thèse de Josipa Cecic-Vidos
04 November 2025
par Isabelle Frapart
November 6, 2025 - Thesis defense by Josipa Cecic-Vidos
04 November 2025
par Isabelle Frapart
2025, November 5 - Thesis defense by Clémence Couton
03 November 2025
par Isabelle Frapart