NMR of Biomolecules: Antimicrobials, Toxins and Metabolites
The team's research contributes to the development of new compounds with antimicrobial or insecticide activity, the identification of new therapeutic targets, or participates in the development of new photoactivatable drugs. Our activities are mainly based on the use of NMR in structural biology and metabolomics. Disulfide Rich Peptides (DRP), AntiMicrobial Peptides (I) and toxins (II) are our privileged subjects of study, and determining their 3D structure is one of the steps in understanding their mechanisms of action. In the CoViD context of recent years, we are also interested in the analysis of spike protein structures for vaccine development (IV). Metabolomics (III), is essentially developed on Drosophila models of human pathologies, to understand which metabolic pathways are impacted by diseases.
I. ANTIMICROBIAL PEPTIDES (Rich in Disulfide Bridges)
Antimicrobial peptides (AMPs), key molecules in the innate immunity of all living organisms, represent one of the most promising alternatives to fight antibiotic resistance, a major public health problem. We are working on a fine understanding of the 3D structures (structural diversity, structure-activity relationships, evolution of structures via phylogeny) and the fine understanding of the mechanisms of action of some promising original AMPs, to help designing new antimicrobial compounds. Avian-double-beta-defensins.
Avian-double-beta-defensins
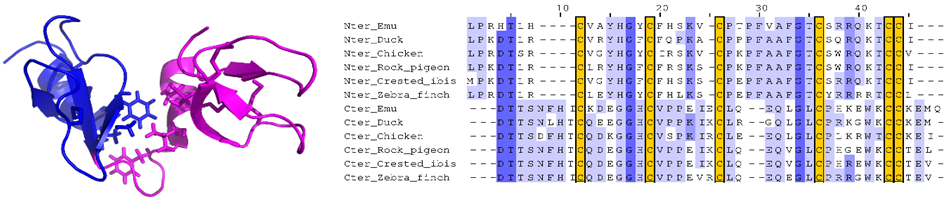 Fig1: We determined the 3D structure of the first avian-double-β-defensin, Gga-AvBD1, from the chicken Gallus gallus, defining a novel 3D fold. We studied several biological activities, including antibacterial, antiparasitic and antiviral, and assessed the contribution of each domain to these activities (Guyot et al. 2020 left). Finally, we showed that the gene coding for this double-defensin, present in all the bird genomes currently available (over 300), does not originate from a duplication-fusion (Guyot et al. 2022 right).
Fig1: We determined the 3D structure of the first avian-double-β-defensin, Gga-AvBD1, from the chicken Gallus gallus, defining a novel 3D fold. We studied several biological activities, including antibacterial, antiparasitic and antiviral, and assessed the contribution of each domain to these activities (Guyot et al. 2020 left). Finally, we showed that the gene coding for this double-defensin, present in all the bird genomes currently available (over 300), does not originate from a duplication-fusion (Guyot et al. 2022 right).
Ancestral oyster defensins 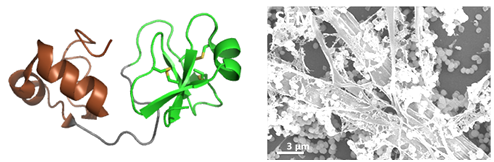 Fig2: Big-defensins are two-domain AMPs, highly diverse in molluscs. We synthesized two big-defensins representative of molecular diversity in the oyster Crassostrea gigas (solid-phase peptide synthesis and native ligation, coll. Equipe Aucagne), and solved their 3D structure (example Cg-BigDef1, left). They include a a β-defensine-like domain (in green) rich in disulfide bridge and hydrophobic N-terminal domain, very preserved, structured in helix-tower-helix (in brown). These defensins are active in salt environment (400 mM NaCl) against a broad spectrum of bacteria. We have shown a synergy of action between the domains, even when they are not covalently linked (Loth et al. 2019) and a remarkable complementarity of their antimicrobial spectrum (DeSanNicolas et al 2022). The hydrophobic domain has also been shown to be indispensable for antimicrobial activity in salty environments, and to govern the self-association of the entire molecule, forming "nanonets" to trap bacteria (Loth et al. 2019, right).
Fig2: Big-defensins are two-domain AMPs, highly diverse in molluscs. We synthesized two big-defensins representative of molecular diversity in the oyster Crassostrea gigas (solid-phase peptide synthesis and native ligation, coll. Equipe Aucagne), and solved their 3D structure (example Cg-BigDef1, left). They include a a β-defensine-like domain (in green) rich in disulfide bridge and hydrophobic N-terminal domain, very preserved, structured in helix-tower-helix (in brown). These defensins are active in salt environment (400 mM NaCl) against a broad spectrum of bacteria. We have shown a synergy of action between the domains, even when they are not covalently linked (Loth et al. 2019) and a remarkable complementarity of their antimicrobial spectrum (DeSanNicolas et al 2022). The hydrophobic domain has also been shown to be indispensable for antimicrobial activity in salty environments, and to govern the self-association of the entire molecule, forming "nanonets" to trap bacteria (Loth et al. 2019, right).
King penguin defensins 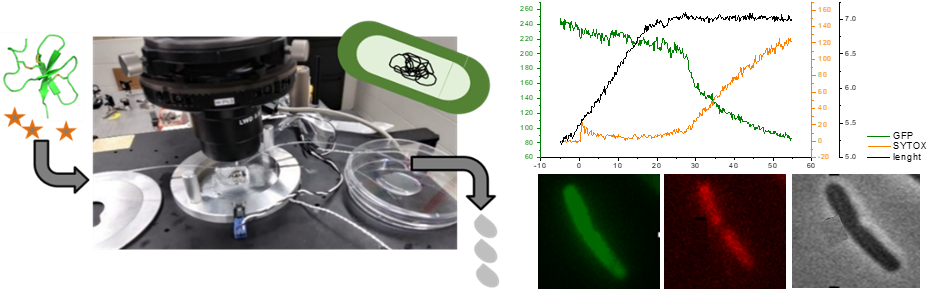 Fig3: An essential first step towards the rational engineering of highly effective antibacterial peptides for clinical use is to understand in detail how nature has optimized AMPs. Data on non-lytic AMPs (i.e. those that do not lyse the bacterial membrane to destroy the bacteria), such as β-defensins, are scarce and therefore remain poorly understood. We study the mechanism of action of defensins, on the example of the king penguin defensin AvBD103b, by performing fluorescence microscopy experiments on living E. coli bacteria that allow us to monitor in real time the slightest permeabilization of their inner and outer membranes (Landon et al. 2022).
Fig3: An essential first step towards the rational engineering of highly effective antibacterial peptides for clinical use is to understand in detail how nature has optimized AMPs. Data on non-lytic AMPs (i.e. those that do not lyse the bacterial membrane to destroy the bacteria), such as β-defensins, are scarce and therefore remain poorly understood. We study the mechanism of action of defensins, on the example of the king penguin defensin AvBD103b, by performing fluorescence microscopy experiments on living E. coli bacteria that allow us to monitor in real time the slightest permeabilization of their inner and outer membranes (Landon et al. 2022).
Other disulfide-rich peptides (Toxines)
Toxins rich in disulfide bridge have strong structural identities with the antimicrobial peptides described above. We study toxins of various origins (plants, ants, spiders, scorpions). We work on the fine understanding of structures (structural diversity, structure-activity relationships, evolution of structures during the evolution of species) to improve understanding of mechanisms of action and participate in the design of the new insecticide compounds. Finally, we are involved in several projects on photo-activatable toxins, which open new therapeutic avenues.
Plant insecticidal toxins 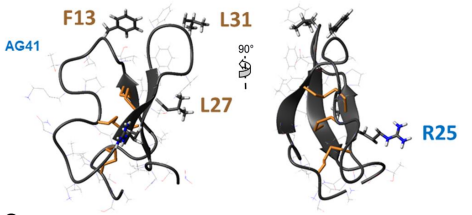 Fig4: Biopesticides are effective alternatives to chemical insecticides. We study the peptide AG41 from the roots of alfalfa, which has exceptional insecticidal activity against several pests such as aphids and weevils. The 3D structure we determined allowed to establish structure-function relationships that explain this high insecticide activity. The key residues for the entomotoxicity of the peptide AG41 are F13, R25, L27 and L31 (Diya et al 2023).
Fig4: Biopesticides are effective alternatives to chemical insecticides. We study the peptide AG41 from the roots of alfalfa, which has exceptional insecticidal activity against several pests such as aphids and weevils. The 3D structure we determined allowed to establish structure-function relationships that explain this high insecticide activity. The key residues for the entomotoxicity of the peptide AG41 are F13, R25, L27 and L31 (Diya et al 2023).
Insecticidal toxins from ant venoms 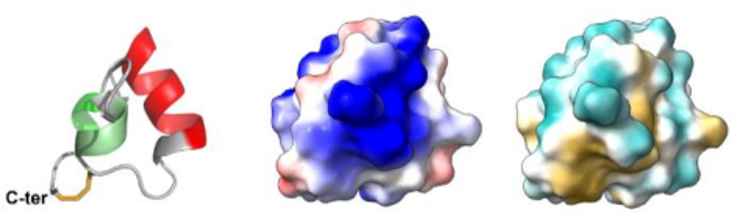 Fig5: Ant venoms, cocktails of toxins optimized to disrupt the physiological systems of prey insects, are a very promising reservoir for the discovery of new insecticides. Among the peptides we studied, we showed that peptide U11 is one of the most paralyzing ever reported from ant venoms against meat blowflies. The U11 peptide has a unique compact helical structure, stabilized by a disulfide bridge (from left to right the 3D structure of the U11 peptide, electrostatic potentials on the surface, hydrophobic potentials on the surface, Barrassé et al 2023).
Fig5: Ant venoms, cocktails of toxins optimized to disrupt the physiological systems of prey insects, are a very promising reservoir for the discovery of new insecticides. Among the peptides we studied, we showed that peptide U11 is one of the most paralyzing ever reported from ant venoms against meat blowflies. The U11 peptide has a unique compact helical structure, stabilized by a disulfide bridge (from left to right the 3D structure of the U11 peptide, electrostatic potentials on the surface, hydrophobic potentials on the surface, Barrassé et al 2023).
Photo-activatable scorpion or spider toxins 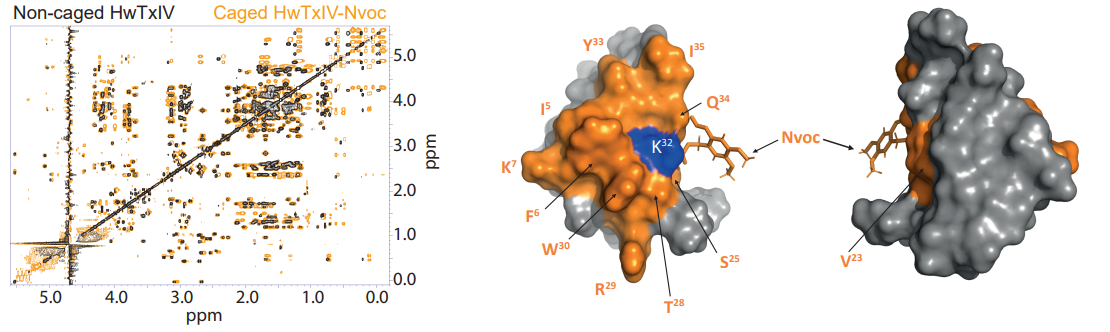 Fig6: The new families of photoactivatable drugs open up new possibilities for therapeutic interventions. In this context, for example, we study toxins rich in disulfide bridges that target the neuronal sodium channels (Montnach et al. 2022) or the ERG cardiac channels (Montnach et al. 2023), and that are rendered photo-activatable by the addition of covalent ligands optimized and cleavable by UV light. Left: example of superimposition of NMR spectra of a spider toxin targeting neuronal ion channels, free or linked to the photo-activatable Nvoc group. Right: 3D structure indicating the area disturbed by the presence of the Nvoc group (Montnach et al. 2022)
Fig6: The new families of photoactivatable drugs open up new possibilities for therapeutic interventions. In this context, for example, we study toxins rich in disulfide bridges that target the neuronal sodium channels (Montnach et al. 2022) or the ERG cardiac channels (Montnach et al. 2023), and that are rendered photo-activatable by the addition of covalent ligands optimized and cleavable by UV light. Left: example of superimposition of NMR spectra of a spider toxin targeting neuronal ion channels, free or linked to the photo-activatable Nvoc group. Right: 3D structure indicating the area disturbed by the presence of the Nvoc group (Montnach et al. 2022)
III. METABOLOMICS
We are developing NMR-based metabolomics analysis, mainly on Drosophila models of human pathologies such as gliomas (Maravat et al. 2021, Bertrand et al. 2023), Huntington's disease (Bertrand et al. 2020) and Parkinson's disease. Other analyses are also being carried out to study different effects, such as that of a therapeutic treatment. The aim of these studies is to identify the metabolic pathways impacted by the diseases in order to better understand them at the molecular level and enable the identification of new therapeutic targets.
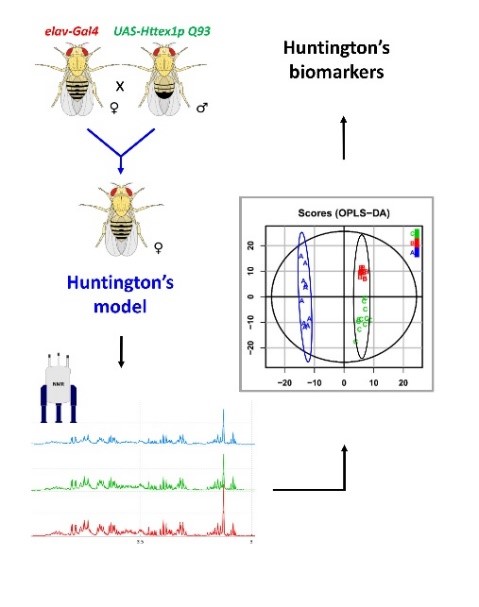
Fig7: Graphical summary of a metabolomics study on the example of Huntington's disease (Bertrand et al. 2020): i) Construction of the Drosophila model carrying Huntington's disease, ii) NMR analyses, iii) Statistical analyses and iv) Identification of disease biomarkers.
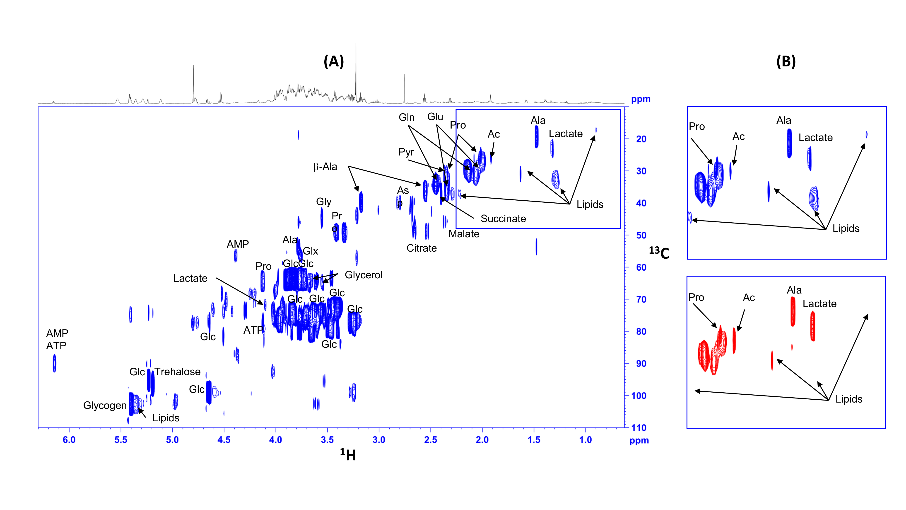
Fig8:13C-HSQC NMR spectra of extracts from control (blue) and glioma (red) Drosophila, fed 13C glucose. .Signals from lipids (clearly visible on enlargements) are considerably reduced in flies carrying gliomas, while lactate is increased (Bertrand et al. 2023).
STRUCTURAL BIOLOGY FOR THE DEVELOPMENT OF SARS-COV-2 VACCINES
Through our expertise in structural biology of proteins, we participated, in collaboration with the Finlay Vaccine Institute of Cuba, in the development of SOBERANA vaccines against COVID19, based on the recombinant protein RBD, SARS-CoV-2 spike protein binding domain on ACE2 receptor.
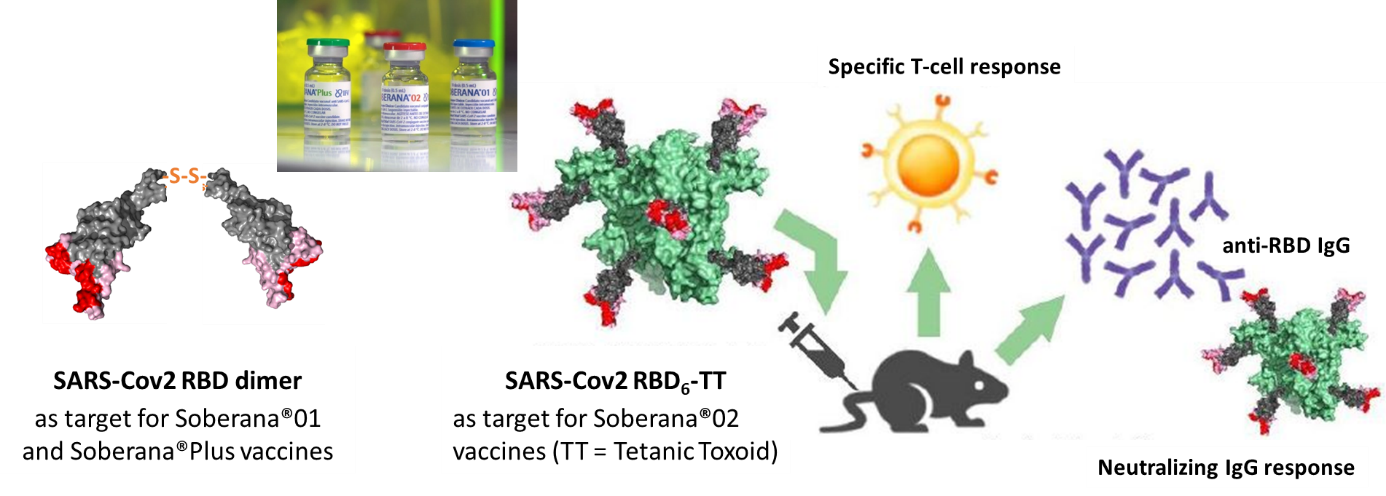
Fig9: The recombinant RBD protein is used in SOBERANA vaccines (Valdes et al ACS Central Sciences 2021) either as a dimer (Santana et al 2022) or conjugated with tetanus anatoxin (Valdes et al ACS ChemBio 2021).
Mains publications (in alphabetical order of first author's name) :
- Barasse V, Jouvensal L, Boy G, Billet A, Ascoet S, Lefranc B, Leprince J, Dejean A, Lacotte V, Rahioui I, Sivignon C, Gaget K, Ribeiro Lopes M, Calevro F, Da Silva P, Loth K, Paquet F, Treilhou M, Bonnafe E and Touchard A. Discovery of an Insect Neuroactive Helix Ring Peptide from Ant Venom. Toxins (Basel). 2023;15.
- Bertrand M, Decoville M, Meudal H, Birman S and Landon C. Metabolomic Nuclear Magnetic Resonance Studies at Presymptomatic and Symptomatic Stages of Huntington’s Disease on a Drosophila Model.Journal of Proteome Research
- Bertrand M, Szeremeta F, Hervouet-Coste N, Sarou-Kanian V, Landon C, Morisset-Lopez S and Decoville M. An adult Drosophila glioma model to highlight metabolic dysfunctions and evaluate the role of the serotonin 5‐HT 7 receptor as a potential therapeutic target. FASEB Journal. 2023;37.
- Diya F, Jouvensal L, Rahioui I, Loth K, Sivignon C, Karaki L, Kfoury L, Rizk F and da Silva P. Residues of Legume AG41 Peptide Crucial to Its Bio-Insecticidal Activity. Biomolecules. 2023;13:446.
- Guyot N, Landon C and Monget P. The Two Domains of the Avian Double-β-Defensin AvBD11 Have Different Ancestors, Common with Potential Monodomain Crocodile and Turtle Defensins. Biology. 2022;11.
- Guyot N, Trapp S, Meudal H, Iochmann S, Silvestre A, Jousset G, Labas V, Reverdiau P, Loth K, Herve V, Aucagne V, Delmas AF, Réhault-Godbert S and Landon C. Structure, function, and evolution of Gga -AvBD11, the archetype of the structural avian-double-β-defensin family. Proceedings of the National Academy of Sciences of the United States of America. 2020;117:337-345.
- Landon C, Zhu Y, Mustafi M, Madinier J-B, Lelièvre D, Aucagne V, Delmas A and Weisshaar JC. Real-Time Fluorescence Microscopy on Living E. coli Sheds New Light on the Antibacterial Effects of the King Penguin beta-Defensin AvBD103b. International Journal of Molecular Sciences. 2022;23:2057.
- Loth K, Vergnes A, Barreto C, Voisin SN, Meudal H, Da Silva J, Bressan A, Belmadi N, Bachere E, Aucagne V, Cazevielle C, Marchandin H, Rosa RD, Bulet P, Touqui L, Delmas AF and Destoumieux-Garzon D. The Ancestral N-Terminal Domain of Big Defensins Drives Bacterially Triggered Assembly into Antimicrobial Nanonets. mBio. 2019;10.
- Maravat M, Bertrand M, Landon C, Fayon F, Morisset-Lopez S, Sarou-Kanian V and Decoville M. Complementary Nuclear Magnetic Resonance-Based Metabolomics Approaches for Glioma Biomarker Identification in a Drosophila melanogaster Model. Journal of Proteome Research. 2021;20:3977-3991.
- Montnach J, Blömer L, Ananda, Lopez L, Filipis L, Meudal H, Lafoux A, Nicolas S, Chu D, Caumes C, Béroud R, Jopling C, Bosmans F, Huchet C, Landon C, de Waard M, Blömer LA and Canepari M. In vivo spatiotemporal control of voltage-gated ion channels by using photoactivatable peptidic toxins. Nature Communications. 2022;13:417.
- Montnach J, Millet H, Persello A, Meudal H, de Waard S, Mesrica P, Ribeiro B, Richard J, Hivonnait A, Tessier A, Lauzier B, Charpentier F, Mangoni M, E, Landon C, Jopling C and de Waard M. Optical Control of Cardiac Rhythm by In Vivo Photoactivation of an ERG Channel Peptide Inhibitor. Circulation Research. 2023;133:535-538.
- Nicolas NdS, Asokan A, Rosa R, Voisin S, Travers M-A, Rocha G, Dantan L, Dorant Y, Mitta G, Petton B, Charrière G, Escoubas J-M, Boulo V, Pouzadoux J, Meudal H, Loth K, Aucagne V, Delmas A, Bulet P, Montagnani C and Destoumieux-Garzón D. Functional Diversification of Oyster Big Defensins Generates Antimicrobial Specificity and Synergy against Members of the Microbiota. Marine drugs. 2022;20:745.
- Santana D, Perez Nicado R, Climent Y, Rodríguez Noda LMM, Sánchez Ramírez B, Perez-Rodriguez S, Rodriguez M, Labrada Regalado C, Hernandez T, Diaz M, Orosa I, Ramirez U, Oliva R, Garrido R, Cardoso F, Landys M, Martinez R, Gonzalez H, Hernandez T, Ochoa-Azze R, Perez J, Enriquez J, Gonzalez N, Infante Y, Espinosa LA, Ramos Y, Gonzalez LJ, Valenzuela C, Casadesus AV, Fernandez B, Rojas G, Pérez-Massón B, Tundidor Y, Bermudez E, Plasencia C, Boggiano T, Ojito E, Chiodo F, Fernandez S, Paquet F, Fang C, Chen G-W, Rivera D, Valdes Y, Garcia-Rivera D and Verez Bencomo V. A COVID-19 vaccine candidate composed of SARS-CoV-2 RBD dimer and Neisseria meningitidis outer membrane vesicles. RSC Chemical Biology. 2022;3:242-249.
- Valdes-Balbin Y, Santana-Mederos D, Quintero L, Fernandez S, Rodriguez L, Sanchez Ramirez B, Perez-Nicado R, Acosta C, Mendez Y, Ricardo MG, Hernandez T, Bergado G, Pi F, Valdes A, Carmenate T, Ramirez U, Oliva R, Soubal JP, Garrido R, Cardoso F, Landys M, Gonzalez H, Farinas M, Enriquez J, Noa E, Suarez A, Fang C, Espinosa LA, Ramos Y, Gonzalez LJ, Climent Y, Rojas G, Relova-Hernandez E, Cabrera Infante Y, Losada SL, Boggiano T, Ojito E, Leon K, Chiodo F, Paquet F, Chen GW, Rivera DG, Garcia-Rivera D and Verez Bencomo V. SARS-CoV-2 RBD-Tetanus Toxoid Conjugate Vaccine Induces a Strong Neutralizing Immunity in Preclinical Studies. ACS Chem Biol. 2021;16:1223-1233.
