Typologie d'actualités: Group RNA Remodeling Factors & Mechanisms
A versatile new approach to seek constitutive or conditional helicase substrates at global scale
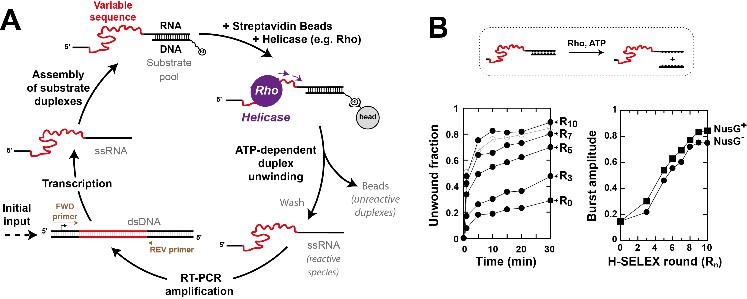
The CNRS Institute of Chemistry has reported this new original screening approach on its website
Référence
Delaleau M., Eveno E., Simon I., Schwartz A & Boudvillain M.
A scalable framework for the discovery of functional helicase substrates and helicase-driven regulatory switches
PNAS 2022

A new mechanism of antiobiotic resistance
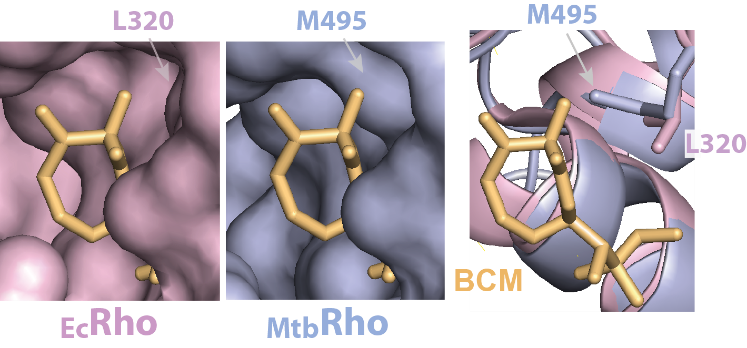
The bacterial Rho factor is a molecular motor that induces genome-wide transcription termination. Rho is essential in many species, including in Mycobacterium tuberculosis where inactivation of the rho gene leads to rapid death. Nevertheless, the Rho factor of M. tuberculosis [MtbRho] displays idiosyncrasies, including resistance to the antibiotic bicyclomycin [BCM], which remain unexplained. To identify the molecular origin of these idiosyncrasies, we solved the structure of MtbRho by cryo-EM at 3.3 Å. This atomic structure notably reveals a leucine → methionine substitution that creates steric hindrance in the binding pockets of BCM, close to the ATPase sites, thereby conferring resistance to BCM at the expanse of molecular motor efficiency. Our work contributes to explain the unusual properties of MtbRho and provides groundwork for the development of new antibiotics.
When a non-conserved protein domain becomes essential
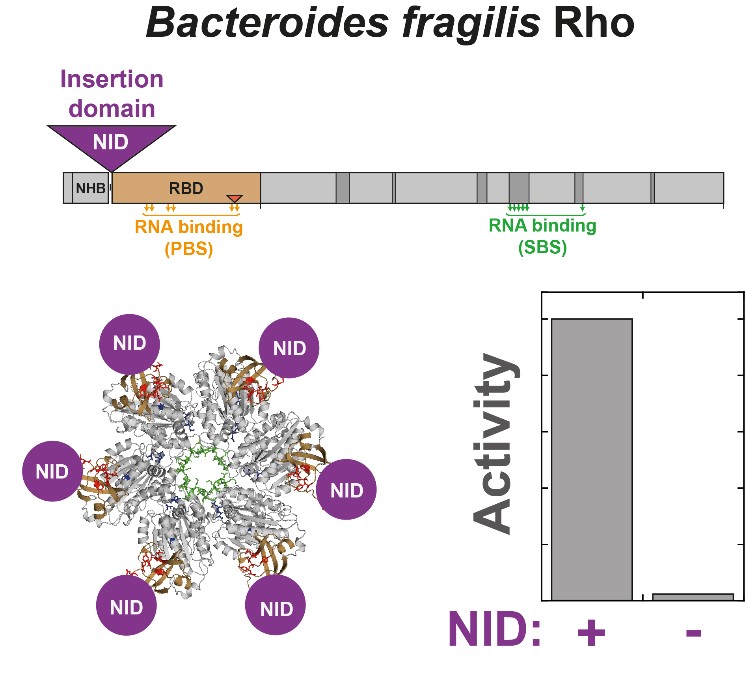
Rho-dependent termination of transcription is a critical regulatory mechanism specific to bacteria. In a subset of species including most Actinobacteria and Bacteroidetes, the Rho factor contains a large, poorly conserved N-terminal insertion domain (NID) of cryptic function. Through the first characterization of an actinobacterial Rho factor containing a very large NID (~40% of total mass), we show that such a non-conserved protein domain can be essential for activity. Without NID, the Rho factor of Bacteroides fragilis (BfRho) indeed cannot induce transcription termination and displays a reduced affinity for RNA. Intriguingly, the presence of a NID in BfRho is not correlated to the lack of residues or motifs deemed essential in NID-less Rho factors from evolutionary distinct species. The NID requirement is probably linked to the coevolution of partner feature(s) such as lineage-specific RNA polymerase domains and/or low G+C content of the B. fragilis transcriptome. Our data thus highlight that ‘essential function’ does not always rhyme with ‘structural conservation’.
Simon I., Delaleau M., Schwartz A., Boudvillain M.
A Large Insertion Domain in the Rho Factor From a Low G + C, Gram-negative Bacterium is Critical for RNA Binding and Transcription Termination Activity
Journal of Molecular Biology (2021) 433 (15) 167060 - Doi : 10.1016/j.jmb.2021.167060
RNA Remodeling Proteins, Methods and Protocols

The second edition of the book “RNA Remodeling Proteins, Methods and Protocols” edited by Marc Boudvillain has just been published.
This volume of the “Methods in Molecular Biology” series compiles new methods to study RNA-protein complexes and the mechanisms leading to their structural remodeling. The volume includes 4 chapters written by CBM researchers and ideally complements the first edition published in 2015.
October 9th, 2020 – External seminar Dr Julien Robert-Paganin, Institut Curie, Paris

Control of bacterial virulence by transcription factors NusG and Rho
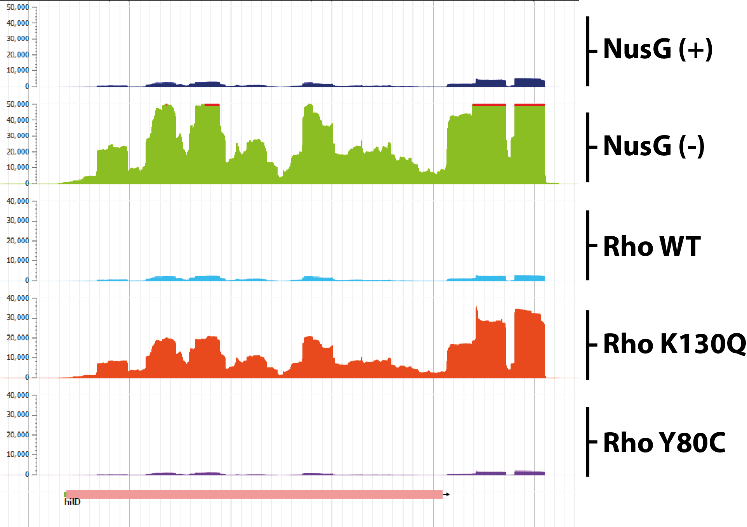
The virulence genes of pathogenic enterobacteria are concentrated in genomic islands acquired by horizontal transfer during evolution. The expression of these genes outside the infection phase is detrimental to the bacterium and is therefore highly regulated. A major regulatory mechanism relies on the histone-like protein H-NS, which binds to AT-rich sites characteristic of horizontally acquired DNA and forms oligomeric structures that inhibit transcription over extended regions. These regions, however, remain exposed to invasive transcription from neighboring regions or to H-NS repression defects. Our data support a model in which the transcription elongation factor NusG “secures” the inhibition of virulence genes by stimulating the activity of the transcription termination factor Rho in regions silenced by H-NS. Remarkably, NusG changes the specificity of the Rho factor, which alone preferentially targets C-rich regions. The perturbation of this NusG/Rho-dependent mechanism in Salmonella has profound physiological consequences, probably because unstopped transcription in H-NS -targeted regions feeds a feed-forward activation cascade leading to the uncontrolled expression of pathogenicity islands and co-regulated loci.



