This group investigates structure-function relationships in RNA-based biological systems. Research is focused on RNA remodelling proteins (helicases, chaperones). The group notably develops sophisticated biochemical and bioinformatics approaches to decrypt the molecular mechanisms of these essential factors.
Research theme
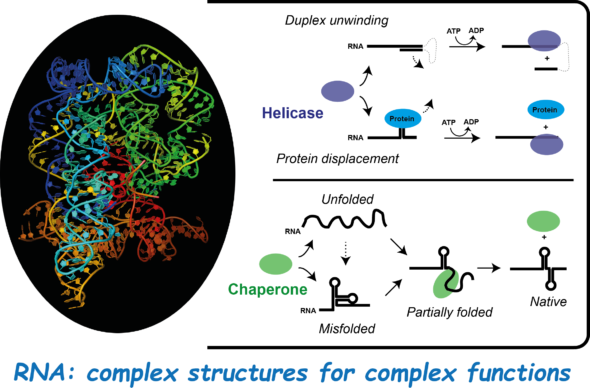
The group explores the mechanisms by which the structures of RNA molecules and ribonucleoproteic (RNP) complexes are remodelled. These structures are often dynamic and can evolve within the cell where they govern the role and fate of RNA species. Two main classes of ubiquitous proteins –RNA chaperones and RNA helicases- shape and remodel the structures of RNA and RNP complexes. The dysfunction of endogenous RNA remodelling proteins or the action of viral or bacterial ones are implicated in diverse pathologies including cancers. We study how RNA helicases and RNA chaperones work and cooperate to drive key physiological functions.
RNA chperones usually disrupt secondary structures upon their binding to RNA and promote the formation of alternative structural motifs and/or complexes. RNA helicases use the energy derived from their NTPase activity to disrupt double-stranded RNA, RNA-DNA heteroduplexes, or RNP complexes. These activities are sometimes linked to directional translocation of the helicase along RNA.
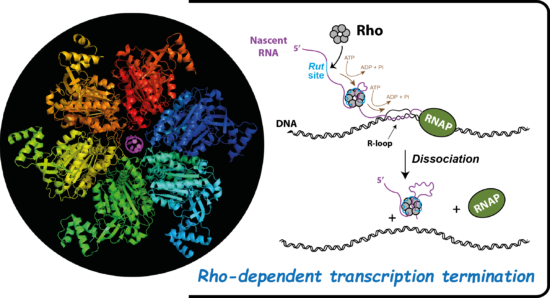
To study RNA remodelling proteins, we use complementary biological, biochemical, and biophysical approaches. We focus our efforts on a complex model system wherein Rho, a ring-shaped RNA helicase, is involved in transcription termination and genome maintenance in bacteria.
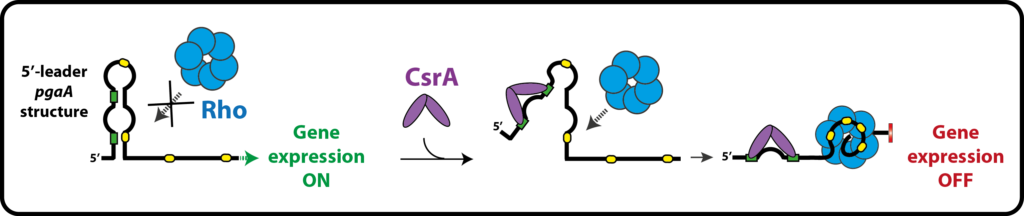
We study the molecular mechanisms governing Rho activity, how the helicase is involved in gene silencing mechanisms implicating non-coding RNAs and RNA chaperones (Hfq, CsrA), the importance of Rho for bacterial pathogenicity and fitness, how the functions and modes of action of Rho are conserved within the biodiversity, etc.
We are also investigating other, distinct RNA helicase specimens. In this way, we hope to understand the basic mechanisms that govern these important enzymes, determine their key common features and differences, and identify factors that contribute to their regulation.
Our long-term goal is to exploit this information for biomedical purposes and for the design of biosensors or inducible gene circuits useful for synthetic biology.
Significant publications :
- Cédric Nadiras, Eric Eveno, Annie Schwartz, Nara Figueroa-Bossi, Marc Boudvillain.
A multivariate prediction model for Rho-dependent termination of transcription.
Nucleic Acids Research (2018) 46, 8245-8260 - Odil Porrua, Marc Boudvillain, Domenico Libri.
Transcription Termination: Variations on Common Themes.
Trends in Genetics (2016) 32, 508-522. - Emilie Soares, Annie Schwartz, Marcello Nollmann, Emmanuel Margeat, Marc Boudvillain.
The RNA-mediated, asymmetric ring regulatory mechanism of the transcription termination Rho helicase decrypted by time-resolved nucleotide analog interference probing (trNAIP).
Nucleic Acids Research (2014) 9270-9284. - Nara Figueroa-Bossi, Annie Schwartz, Benoit Gillemardet, François D’Heygère, Lionello Bossi, Marc Boudvillain
RNA remodeling by bacterial global regulator CsrA promotes Rho-dependent transcription termination
Genes & Development (2014) 28, 1239-51. - Rachid Menouni, Stéphanie Champ, Léon Espinosa, Marc Boudvillain, Mireille Ansaldi
Transcription termination controls prophage maintenance in Escherichia coli genomes
Proc. Natl. Acad. Sci USA (2013) 110, 14414-14419.