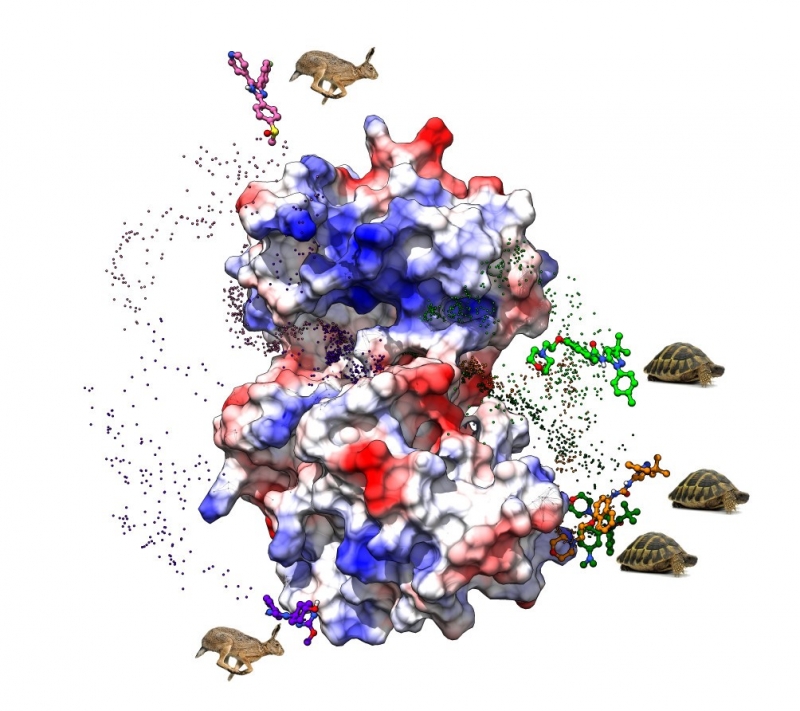
Chemists from the Institute of Organic and Analytical Chemistry (ICOA, CNRS / University of Orléans) and the Center for Molecular Biophysics (CBM, CNRS) propose a new in silico model, which describes the duration of interactions between a molecule and its biological target. Published in the Journal of Chemical Information and Modeling, this work has successfully predicted effects on a protein linked to certain cancers and helps to reduce doses and thus toxicity.
Catégorie d'actualités: significant publications
Antimicrobial peptides: an atypical double-domain avian defensin, specifically found in eggs, reveals multiple roles in protection of the embryo
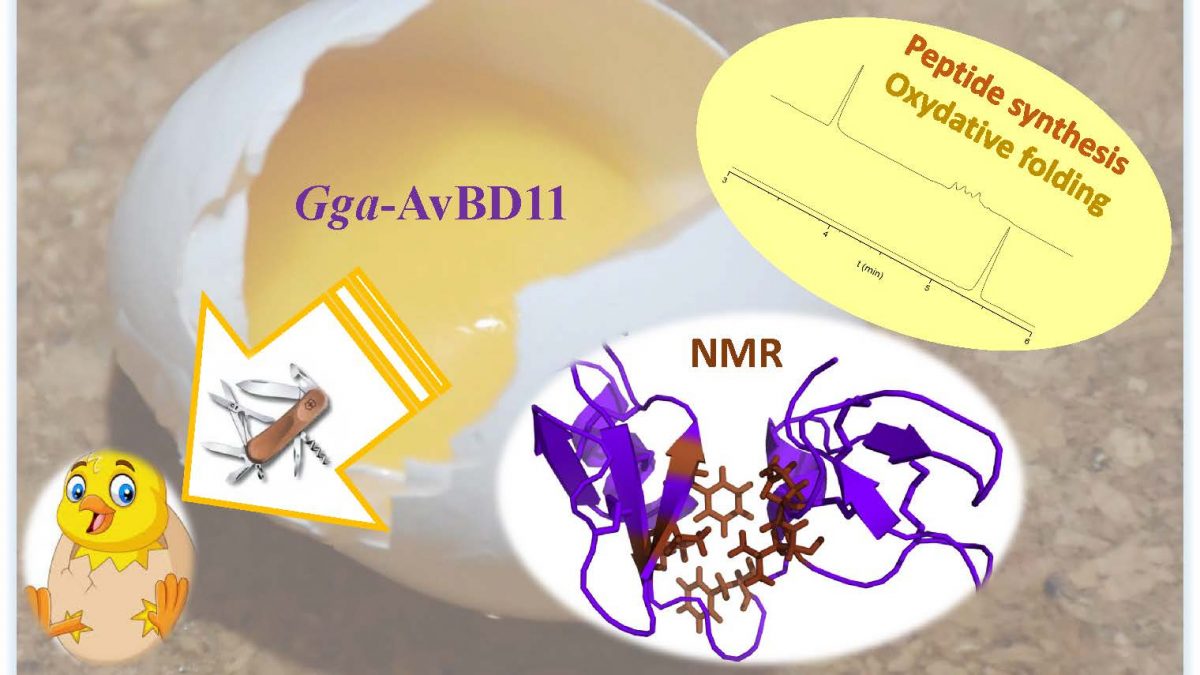
Gga-AvBD11, the avian β-defensin 11 of the common chicken Gallus gallus (Gga-AvBD11), is egg-specific, and represents the sole double-sized defensin (9.3 kDa) among the 14 AvBDs reported in the chicken species. The appearance of such a double-domain protein during evolution could be driven either by its increased biological potency compared to a single domain molecule, and/or by the necessity to acquire new functions carried only by the full-length protein. To assess the contribution of the two domains, we chemically synthesized them. We determined the 3D NMR structure each domain, and the structure of the compact full-length Gga-AvBD11, composed of two packed β-defensin domains. There is no reference for such a double-β-defensin in structural databanks. Thus, AvBD11 is the archetype of a new structural family, which we named avian-double-β-defensins (Av-DBD).
Its high sequence conservation among birds suggests its essential roles in the avian egg. In collaboration with several teams (Nouzilly and Tours, France), we showed that Gga-AvBD11 displays antimicrobial activities against Gram+ and Gram- bacterial pathogens, the avian protozoan Eimeria tenella and avian influenza virus (H1N1). It also shows cytotoxic and anti-invasive activities, suggesting that it may be involved in the (re-)modeling of embryonic tissues. Our results point to a critical importance of the cationic N-ter domain in mediating antibacterial, antiparasitic and anti-invasive activities, with the C-ter domain potentiating the two latter activities. Strikingly, antiviral activity in infected chicken cells requires the full-length protein.
The benefit for the avian species of possessing a double-sized defensin is a fascinating question. In order to better understand the structure-activity and phylogenetic relationships of AvBD11s family, we are currently studying other AvBD11 proteins (SAPhyR-11 project grant from the Région Centre Val de Loire).
This work was funded by the MUSE (Grant no. 2014-00094512) and SAPhyR-11 (Grant no. 2017-119983) project grants from the Région Centre-Val de Loire.
Structure, function, and evolution of Gga-AvBD11, the archetype of the structural avian-double-β-defensin family
Nicolas Guyot, Hervé Meudal, Sascha Trapp, Sophie Iochmann, Anne Silvestre, Guillaume Jousset, Valérie Labas, Pascale Reverdiau, Karine Loth, Virginie Hervé, Vincent Aucagne, Agnès F. Delmas, Sophie Rehault-Godbert, and Céline Landon
PNAS January 7, 2020 117 (1) 337-345; first published December 23, 2019 https://doi.org/10.1073/pnas.1912941117
Antimicrobial peptides: How peptide chemistry and NMR shed light on the antimicrobial activity of big defensins
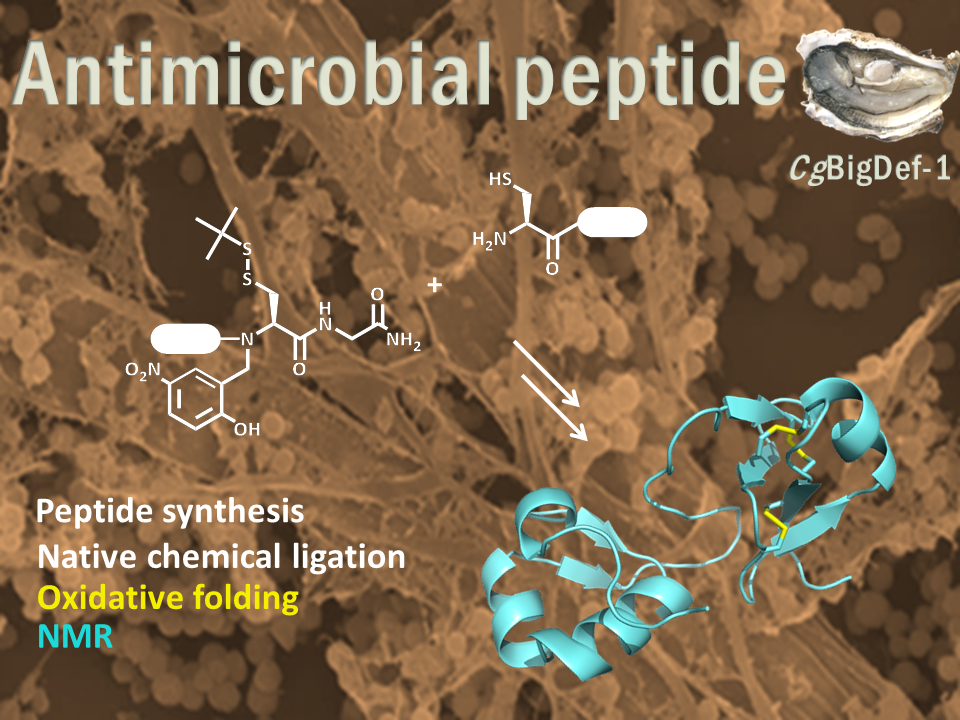
Big defensins, ancestors of β-defensins, are composed of a β-defensin-like C-terminal domain and a hydrophobic ancestral N-terminal domain.
This unique structure is found in a limited number of phylogenetically distant species, mostly living in marine environments. Using solid phase peptide chemistry and native chemical ligation, we produced the oyster Crassostrea gigas BigDef1 (Cg-BigDef1) and its separate domains and characterized their 3D structure by NMR. Cg-BigDef1 showed salt-stable and broad-range bactericidal activity, including against multidrug-resistant clinical isolates of S. aureus. We found that the ancestral N-terminal domain confers salt-stable antimicrobial activity to the β-defensin-like domain, which is otherwise inactive. Moreover, upon contact with bacteria, the N-terminal domain drives Cg-BigDef1 assembly into nanonets that entrap and kill bacteria. We speculate that the hydrophobic N-terminal domain of big defensins has been retained in marine phyla to confer salt-stable interactions with bacterial membranes in environments where electrostatic interactions are impaired.
Those remarkable properties open the way to future drug developments when physiological salt concentrations inhibit the antimicrobial activity of vertebrate β-defensins (ANR MOSAR-Def 2019-2023).
Many thanks to D. Destoumieux-Garzón for collaboration, to “Vaincre La Mucovidose“ and CNRS PEPS X-life” for funding.
Control of bacterial virulence by transcription factors NusG and Rho
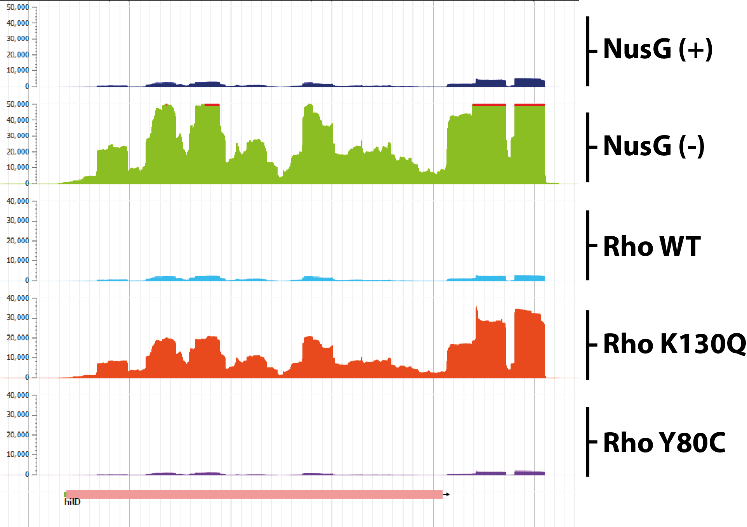
The virulence genes of pathogenic enterobacteria are concentrated in genomic islands acquired by horizontal transfer during evolution. The expression of these genes outside the infection phase is detrimental to the bacterium and is therefore highly regulated. A major regulatory mechanism relies on the histone-like protein H-NS, which binds to AT-rich sites characteristic of horizontally acquired DNA and forms oligomeric structures that inhibit transcription over extended regions. These regions, however, remain exposed to invasive transcription from neighboring regions or to H-NS repression defects. Our data support a model in which the transcription elongation factor NusG “secures” the inhibition of virulence genes by stimulating the activity of the transcription termination factor Rho in regions silenced by H-NS. Remarkably, NusG changes the specificity of the Rho factor, which alone preferentially targets C-rich regions. The perturbation of this NusG/Rho-dependent mechanism in Salmonella has profound physiological consequences, probably because unstopped transcription in H-NS -targeted regions feeds a feed-forward activation cascade leading to the uncontrolled expression of pathogenicity islands and co-regulated loci.
Everything you’ve always wanted to know about protein/protein interactions and kinase activity assays is in a paper published in JOVE.
"Cell signalling" group led by the Dr H. Bénédetti has published a video paper in JOVE, Journal of Visualized Experiments, a peer-reviewed scientific journal that publishes experimental methods in video format.
This paper belongs to the Method section focusing on basic technics in Biochemistry.
Different experiments are detailled:
- Transient cell transfections
- Protein extraction
- Study of protein/protein interaction by co-immunoprecipitation experiments
- Study of kinase activity by g32P-[ATP] labeling or by using phospho-specific antibodies
All these different technics are illustrated with data obtained by the group on LIMK2-1, a new protein the team has just pointed out and characterized. LIMK2-1 protein does exist, and it is very atypical in the way it regulates actin cytoskeleton remodeling.
Extraterrestrial organic matter that fell on Earth 3.3 billion years ago

The meteorites bring it regularly, but it has left no trace of it. It has been identified and analyzed by researchers from the CNRS, Chimie ParisTech, the Universities of Tours and Lille. Their publication, in the journal Geochimica and Cosmochimica Acta, offers a first model to distinguish these molecules from elsewhere from those produced on Earth.
References of the article:
D. Gourier et al.
Extraterrestrial organic matter preserved in 3.33 Ga sediments from Barberton, South Africa.
Geochimica and Cosmochimica Acta - May 2019 - DOI: 10.1016 / j.gca.2019.05.009
A new interpretation of neutron scattering spectra
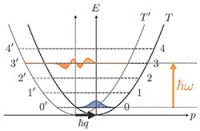
The article presents a new interpretation of neutron scattering spectra by molecular systems that has much in common with the Franck-Condon theory describing the vibrational transitions in a molecule after absorption or emission of a photon. The principal elements are the quantum probabilities for the transitions between the energy levels of the studied system, which are induced by diffusion of a neutron. In this case, the fundamental concept of "energy landscapes", which was introduced by Hans Frauenfelder to describe the internal dynamics of proteins in terms of "jumps" between the minima of their (free) internal energy, can be integrated in the analysis of neutron scattering spectra by complex systems in general. The theory also provides an intuitive physical interpretation of Van Hove's correlation functions in the quantum regime, as well as their classical limit, which is usually considered in the analysis of quasi-elastic spectra of neutrons from proteins and other complex molecular systems.