Typologie d'actualités: Group DNA repair: Structure, Function and Dynamic
Protein filaments in the regulation of gene expression
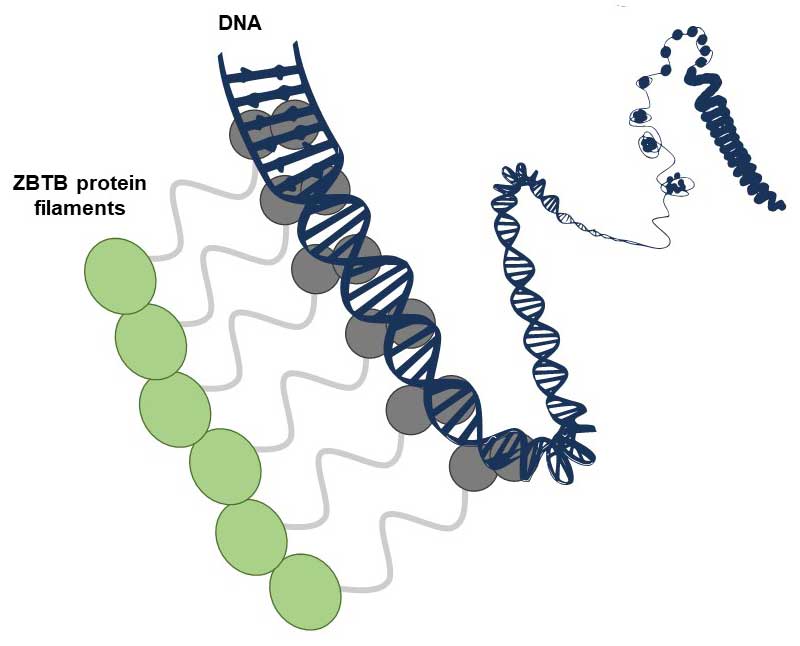
Although every cell in our body contains the same genetic information, cells differ in the way they use it, a process known as “gene expression”. The regulation of gene expression is orchestrated by proteins called transcription factors, which bind to specific sequences within DNA. Transcription factors are traditionally thought to operate mainly as single molecules or dimers.
The article by Mance et al. reveals that several transcription factors of the family known as ZBTB, present in humans and other animals, have the capacity to form non-covalent filamentous structures composed of numerous identical copies of proteins arranged in a chain. At the molecular level, such structures could offer significant advantages for binding to DNA, which is itself an elongated molecule containing numerous repeated sequences. A few examples of filament-forming transcription factors had already been reported, but this study extends the concept to a large family of this protein with important functions. The study - which combines structural, biophysical and functional analysis carried out in vitro and in cells - was carried out by the "Post-translational modifications and DNA repair" team at the CBM and their collaborators in Orléans, Rennes and Marseille, including the "Functional mass spectrometry of molecular assemblies" team also at the CBM.
The findings from this research, together with a complementary study by groups of Benjamin Ebert and Eric Fischer from Dana-Farber Cancer Institute at Harvard (published back-to-back in the same issue of Molecular Cell), challenge the traditional view of transcription factor functionality.
In cells, ZBTB proteins are regulated through a process called SUMOylation, where a small tag called SUMO is added to them, changing how they function. Studies on ZBTB proteins undertaken in Orléans, during which the filamentous structures were discovered, are part of the "SUMOwriteNread" project funded by the European Union (ERC grant no 101078837). Researchers are currently investigating the interplay between the ability to form filaments and SUMO tagging to understand the complex reality of gene expression regulation.
This research was reported by CNRS Chimie on its website.
Dynamic BTB-domain filaments promote clustering of ZBTB proteins.
Lucija Mance, Nicolas Bigot, Edison Zhamungui Sánchez, Franck Coste, Natalia Martín-González, Siham Zentout, Marin Biliškov, Zofia Pukało, Aanchal Mishra, Catherine Chapuis, Ana-Andreea Arteni, Axelle Lateur, Stéphane Goffinont, Virginie Gaudon, Ibtissam Talhaoui, Ignacio Casuso, Martine Beaufour, Norbert Garnier, Franck Artzner, Martine Cadene, Sébastien Huet, Bertrand Castaing & Marcin Józef Suskiewicz
Molecular Cell 2024
https://doi.org/10.1016/j.molcel.2024.05.029
2024, April 19 – Seminar of Dimitar Angelov

Breast cancer: towards early diagnosis by imaging
In vivo imaging of metastatic breast cancer tumors at very early stages is about to become possible. A team of chemists and biologists from the Center for Molecular Biophysics (CNRS) has indeed developed a new magnetic resonance imaging (MRI) probe which has a selective affinity for an emerging biomarker of metastatic breast cancer: Netrin- 1. Find out more on the Cnrs Chimie website.
See more and Cnrs Chmie website.
Reference
Peptide-Conjugated MRI Probe Targeted to Netrin-1, a Novel Metastatic Breast Cancer Biomarker
Clémentine Moreau, Tea Lukačević, Agnès Pallier, Julien Sobilo, Samia Aci-Sèche, Norbert Garnier, Sandra Même, Éva Tóth & Sara Lacerda
Bioconjugate Chemistry 2024
https://doi.org/10.1021/acs.bioconjchem.3c00558
Comprehensive review about the “logic of protein modifications”
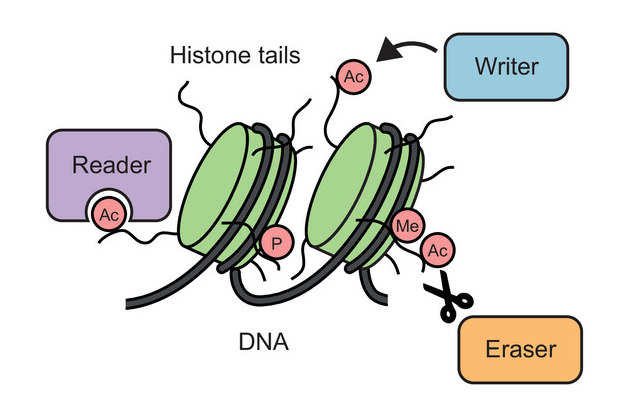
Among the main functional building blocks of living cells are proteins, small “molecular machines” produced by the cell according to the information encoded in genes. Each protein has its characteristic chemical composition which defines its structure and function.
In some circumstances, the chemical composition of a protein can be changed in an enzymatic process known as post-translational modification (PTM), whereby additional chemical groups are covalently attached to the protein. PTMs are used by the cell as a regulatory mechanism to control protein function. The addition of new chemical groups – which can come in different shapes and sizes, ranging from small groups, through sugars and lipids, to small proteins – changes the structure and interactions of a protein and can impact almost any aspect of its function.
Marcin Suskiewicz, a structural biologist and biochemist from the CBM, has devoted many years to studying various types of protein PTMs and currently supervises a project devoted to one particular type of PTMs, protein SUMOylation.
In the review published in the journal BioEssays, he reviews the history of the research into protein PTMs as well as various facets of this phenomenon, including the underlying chemical principles, molecular mechanisms, and evolution.
The review combines an introduction to the field with an overview of the recent literature and new ideas and hypotheses.
References:
The logic of protein post-translational modifications (PTMs): Chemistry, mechanisms and evolution of protein regulation through covalent attachments
Marcin Suskiewicz
BioEssays
First published:21 January 2024
https://doi-org.insb.bib.cnrs.fr/10.1002/bies.202300178
2023.12.08 – Seminar of Ireneusz LITWIN

A review on ADP-ribosylation appeared in the journal Cell
ADP-ribosylation is a biochemical reaction in which the ADP-ribose group from NAD+ becomes covalently attached to various substrates. As such, ADP-ribosylation represents a ubiquitous modification of proteins and other biomolecules (e.g., nucleic acids). Catalysed by a range of specific enzymes, the most important of which in humans is PARP1, ADP-ribosylation serves as a regulatory mechanism influencing a wide array of cellular processes in all domains of life. This new review, published in the authoritative Leading Edge series of reviews of the journal Cell, covers the state of the art on this subject spanning structural biology, biochemistry, cell biology, and the clinical facets of ADP-ribosylation. In addition to Marcin Suskiewicz from the CBM as the first author, the review was co-authored by Ivan Ahel and members of his group at the University of Oxford.
Suskiewicz M., Prokhlrova E., Rack J.G.M., Ahel I.
ADP-ribosylation from molecular mechanisms to therapeutic implications
Cell Review, Volume 186, Issue 21, pages 4475-4495, October 12, 2023 - doi: 10.1016/j.cell.2023.08.03



