|
Gadolinium (Gd3+) complexes have been used as MRI contrast agents for 35 years, but recently the safety of some was questioned. The replacement of Gd3+ by manganese (Mn2+), a biogenic metal, would enable safer complexes.
Nevertheless, the Mn2+ has to be chelated by complexes exhibiting high thermodynamic stability and kinetic inertness (to guarantee that the Mn is not released in vivo) and with a water molecule directly coordinated to the metal, essential for a good MRI efficiency. Combining these two properties is a chemistry challenge.
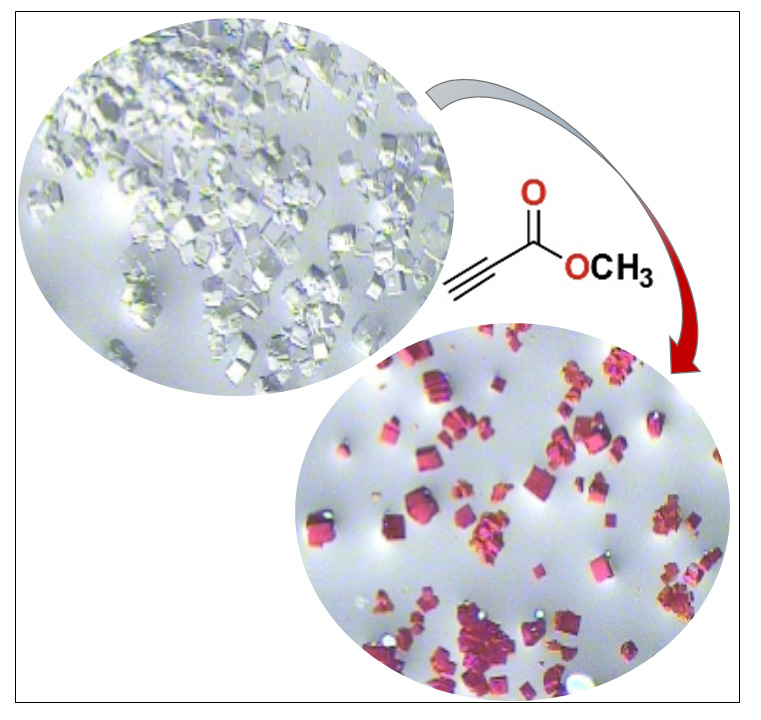
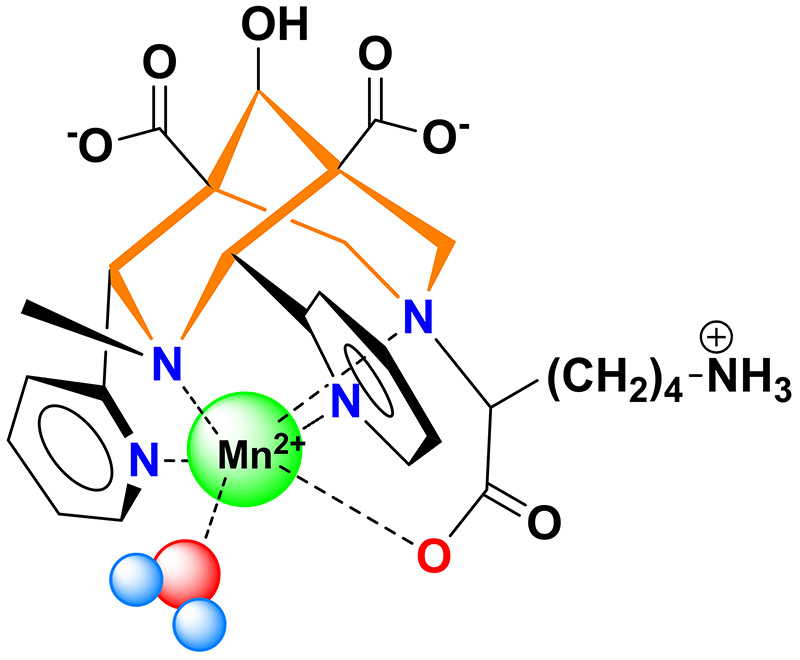
The “Metal complexes and MRI” team of CBM and their collaborators from IPHC (Strasbourg) have synthesized and studied a bispidine ligand, a molecule which cavity is well adapted for Mn2+ complexation. This Mn2+ complex has an excellent kinetic inertness and its MRI efficiency was validated by preclinical studies.
52Mn is an emergent radionuclide for positron emission tomography (PET). Mn2+ is the only metal enabling both MRI and PET imaging. The use of 52Mn is nevertheless limited by its low availability and lack of appropriate ligand.
For the first time in France, 52Mn was produced at the Orléans’ cyclotron, and 52Mn-bispidine was successfully obtained.
Overall, bispidine is a very promising ligand for the Mn2+ complexation, for MRI and PET. Due to its outstanding kinetic inertness, in vivo use of Mn2+ without toxicity risk can be anticipated.
See the news on the website of the CNRS Institute for Chemistry.
Eva Toth, Daouda Ndiaye, Maryame Sy, Agnès Pallier, Sandra Même, Isidro de Silva, Sara Lacerda, Aline M. Nonat, Loïc J. Charbonnière Unprecedented kinetic inertness for a Mn2+‐bispidine chelate: a novel structural entry for Mn2+‐based imaging agents - Angewandte Chemie, 2020, https://doi.org/10.1002/anie.202003685
|