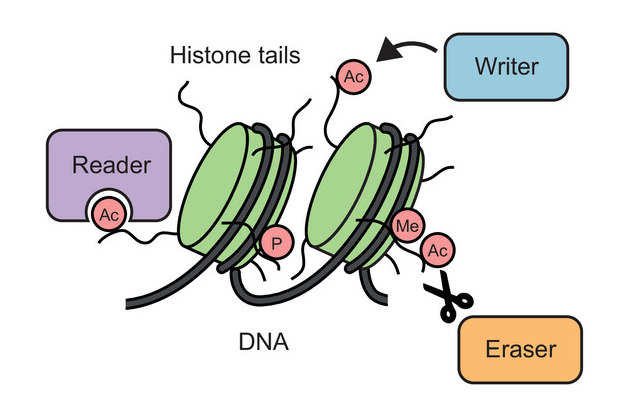
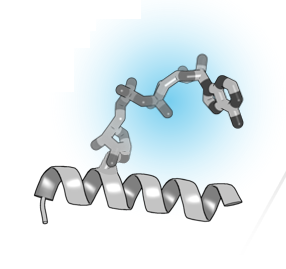
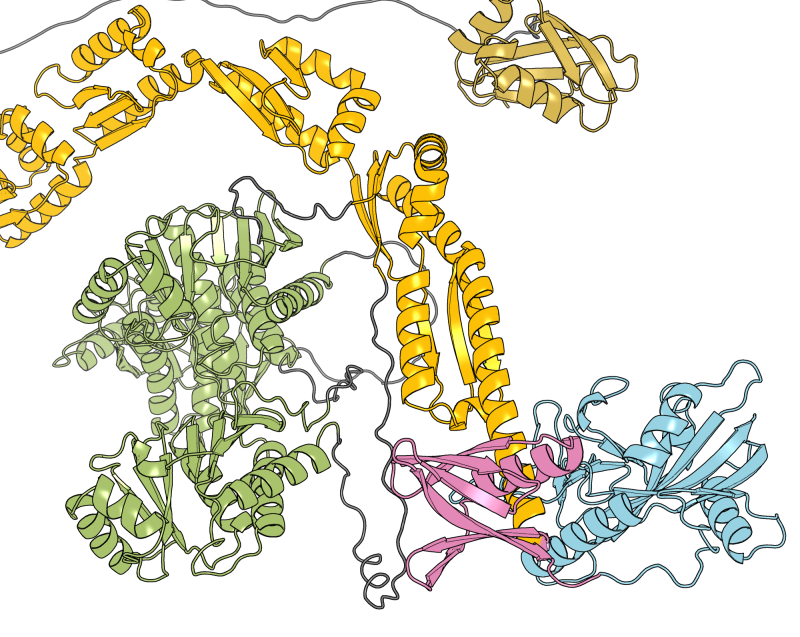
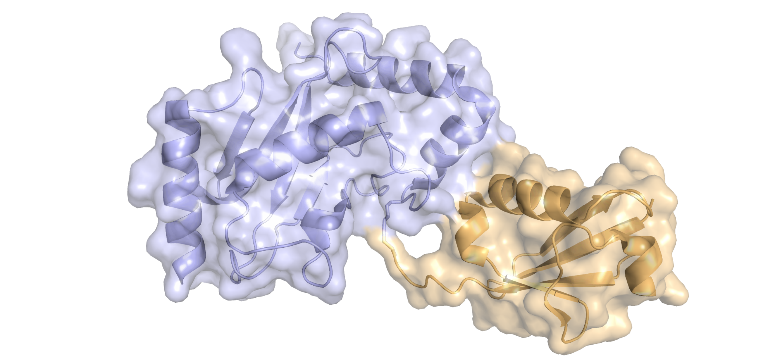
Among the main functional building blocks of living cells are proteins, small “molecular machines” produced by the cell according to the information encoded in genes. Each protein has its characteristic chemical composition which defines its structure and function.
In some circumstances, the chemical composition of a protein can be changed in an enzymatic process known as post-translational modification (PTM), whereby additional chemical groups are covalently attached to the protein. PTMs are used by the cell as a regulatory mechanism to control protein function. The addition of new chemical groups – which can come in different shapes and sizes, ranging from small groups, through sugars and lipids, to small proteins – changes the structure and interactions of a protein and can impact almost any aspect of its function.
Marcin Suskiewicz, a structural biologist and biochemist from the CBM, has devoted many years to studying various types of protein PTMs and currently supervises a project devoted to one particular type of PTMs, protein SUMOylation.
In the review published in the journal BioEssays, he reviews the history of the research into protein PTMs as well as various facets of this phenomenon, including the underlying chemical principles, molecular mechanisms, and evolution.
The review combines an introduction to the field with an overview of the recent literature and new ideas and hypotheses.
References:
The logic of protein post-translational modifications (PTMs): Chemistry, mechanisms and evolution of protein regulation through covalent attachments
Marcin Suskiewicz
BioEssays
First published:21 January 2024
https://doi-org.insb.bib.cnrs.fr/10.1002/bies.202300178