The use of different original NMR methods (MRI, 1H HR-MAS, 2D liquid NMR) made it possible to characterize a glioma model established in adult Drosophila and to reveal the therapeutic potential of a serotonin receptor for the treatment of these cancers.
Read the "Actualité chimique" article n° 492, 2024, February
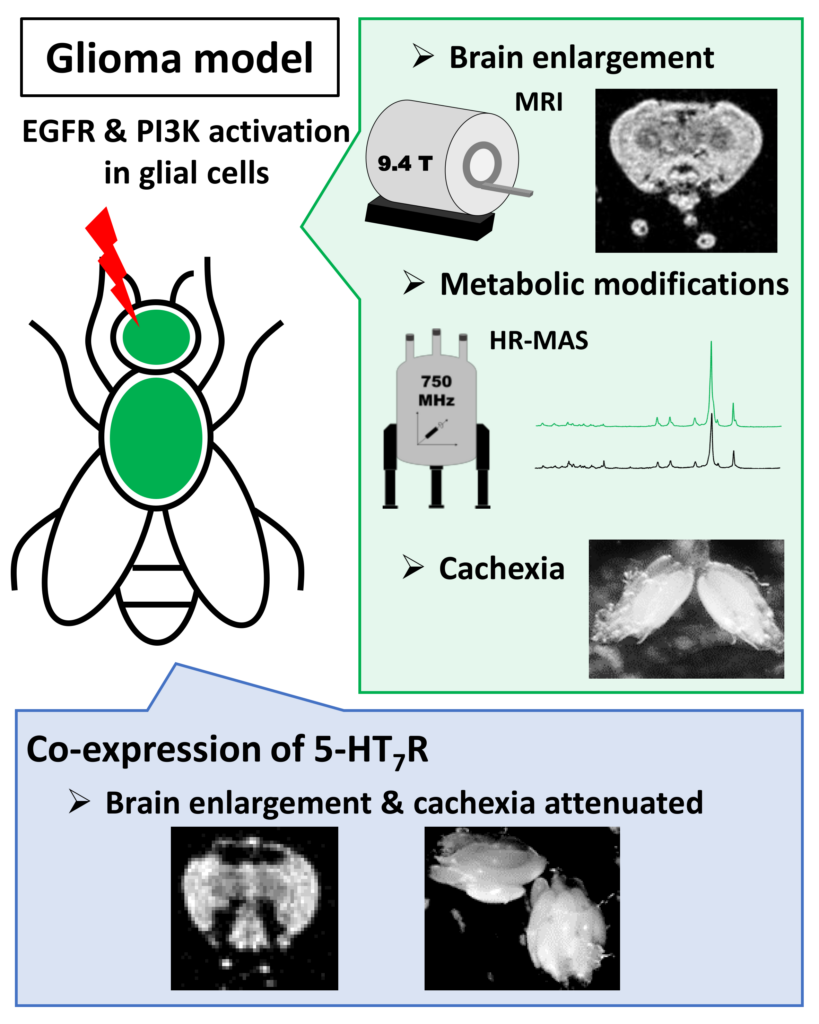
Gliomas account for 50% of brain cancers and therefore constitute the most common brain tumors. Molecular alterations involved in adult gliomas have been identified and mainly affect tyrosine kinase receptors with amplification and/or mutation of the epidermal growth factor receptor (EGFR) and its associated signaling pathways. Several targeted therapies have been developed but current treatments remain ineffective for glioblastomas, the most severe forms. Thus, it is a priority to identify new pharmacological targets. Here, we used a Drosophila glioma model in adult, to characterize metabolic disturbances associated with glioma and assess the consequences of the serotonin 5-HT7 receptor expression on glioma development. First, by using in vivo Magnetic Resonance Imaging, we have shown that expression of the constitutively active forms of EGFR and PI3K in adult glial cells induced enlargement of brains. Then, we explored altered cellular metabolism by using High-Resolution Magic Angle Spinning NMR and 1H-13C Heteronuclear Single Quantum Coherence solution state. Discriminant metabolites identified highlight rewiring of metabolic pathways in glioma, and associated cachexia phenotypes. Finally, the expression of 5-HT7R in this model attenuates phenotypes associated with glioma development (brain enlargement and cachexia).
Article :
An adult Drosophila glioma model to highlight metabolic dysfunctions and evaluate the role of the serotonin 5-HT7 receptor as a potential therapeutic target.
Bertrand M, Szeremeta F, Hervouet‐Coste N, Sarou-Kanian V, Landon C, Morisset-Lopez S, Decoville M
The FASEB Journal. 2023 37:e23230. doi:10.1096/fj.202300783RR