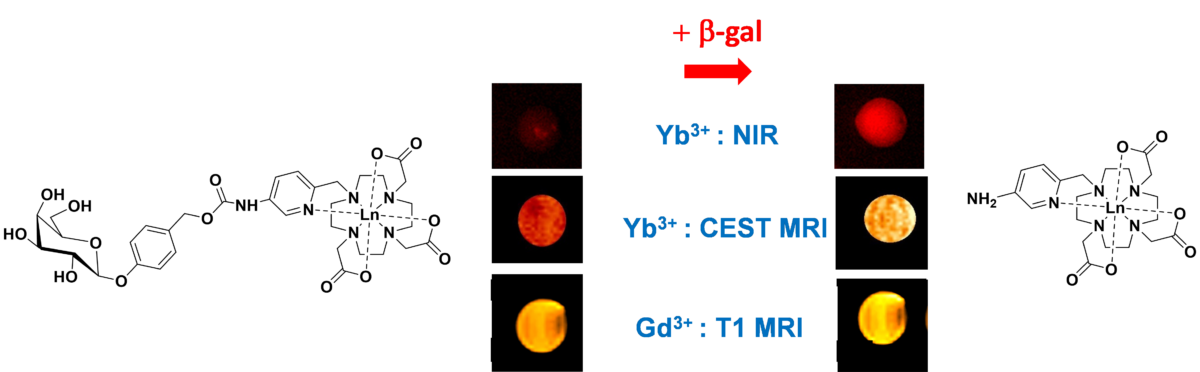
The imaging visualization of active enzymes is of primary importance in biology.
In a collaborative effort between the Centre of Molecular Biophysics (CBM) and the Institute of Chemistry of Natural Substances (ICSN) in Gif sur Yvette, CBM researchers have designed Ln3+ complexes that provide enzyme-mediated changes in NIR luminescence, as well as in Chemical Exchange Saturation Transfer (CEST) and classical T1-weighted MRI, depending on the Ln3+ used. They have demonstrated the successful monitoring of b-galactosidase activity over time in NIR luminescence and CEST MR imaging in phantoms containing the Yb-complex, and in T1 MRI when using the Gd-analogue. A further great advantage of their probe design is its high versatility, as there are a large number of enzymatically cleavable groups that could be attached to the same core, thus creating probes for other important enzyme targets.
Reference : Rémy Jouclas, Sophie Laine, Svetlana V. Eliseeva, Jérémie Mandel, Frédéric Szeremeta, Pascal Retailleau, Jiefang He, Jean-Francois Gallard, Agnès Pallier, Célia S. Bonnet, Stéphane Petoud, Philippe Durand, Éva Tóth
Lanthanide-Based Probes for Imaging Detection of Enzyme Activities by NIR Luminescence, T1- and ParaCEST MRI
Angew. Chem. Int. Ed. 2024, https://onlinelibrary.wiley.com/doi/10.1002/anie.202317728