Physiological characterization of life in Engineered Living Materials by confocal microscopy at single cell resolution.
The CNRS Institute of Chemistry reported this remarkable research on its site. See the article
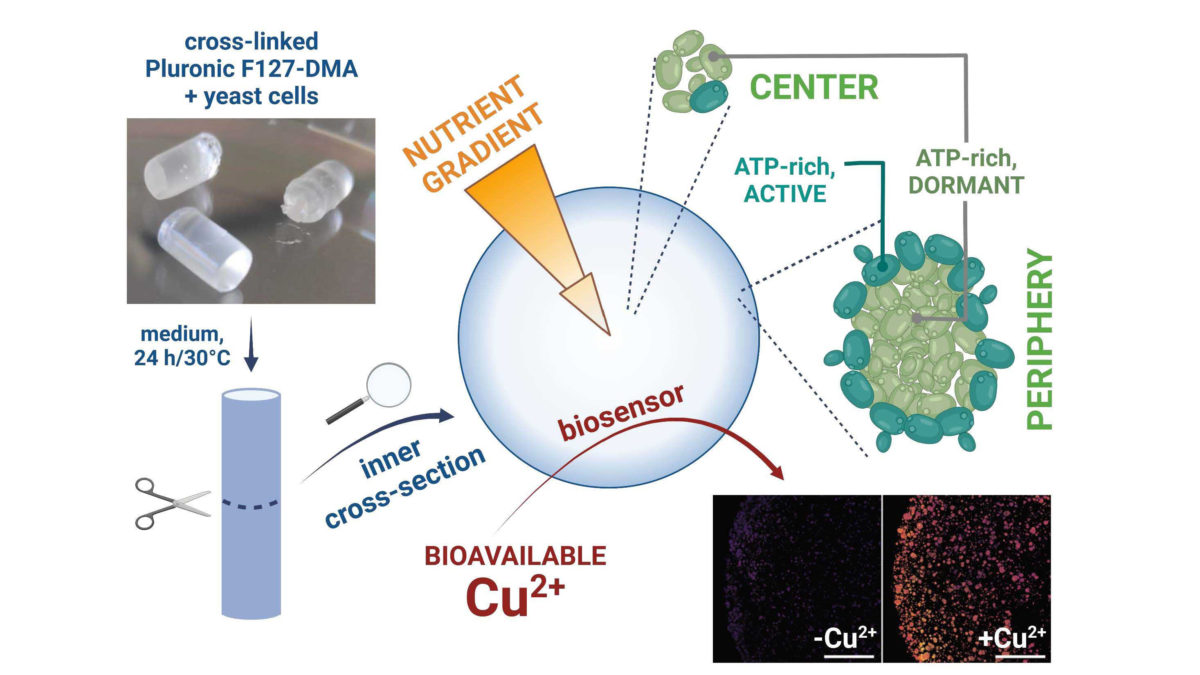
Engineered Living Materials (ELMs) combine living cells with non-living scaffolds to get life-like characteristics, such as biosensing, growth, and self-repair. Some ELMs are 3D-printed, and called bio-ink. For ELMs to be functional, cells in ELMs has to remain alive and active. However, currently, microorganism physiology in ELMs is still elusive and restrict their use.
Researchers of the team "Cell signalling and neurofibromatosis" reconstituted such bioprinted ELMs by associating the yeast Saccharomyces cerevisiae with the hydrogel Pluronic F-127. Theydeveloped genetically engineered yeast by integrating fluorescent gene whose expression is correlated to a physiological parameter: ATP concentration (metabolism), intracellular pH (growth phase), morphology … These engineered and ratiometric biosensors are effective and allow to assess yeast physiological status in ELM directly in situ by confocal microscopy at single cell scale level. They constitute a valuable tool easy to adapt to any other system by associating them to other materials to evaluate their biocompatibility.
Furthermore, the researchers tested their recently developed copper biosensor embedded into this hydrogel F-127, and showed it is fully functional into this ELM. Yeast biosensor association with hydrogel provides several very interesting advantages such as protecting yeast from contaminations and supplying them with nutrients.
This work allows to establish the proof of concept that F127 associated with engineered yeast S. cerevisiae is a promising ELM in order to develop easy to use whole-cell biosensors able to detect copper directly on samples collected in the environment.
Bojan Žunar B., Ito T., Mosrin C., Sugahara Y., Bénédetti H., Guégan R. and Vallée B.
Confocal imaging of biomarkers at a single-cell resolution: quantifying 'living' in 3D-printable engineered living material based on Pluronic F-127 and yeast Saccharomyces cerevisiae.
Biomater Res 26, 85 (2022). https://doi.org/10.1186/s40824-022-00337-8